Type II Tools
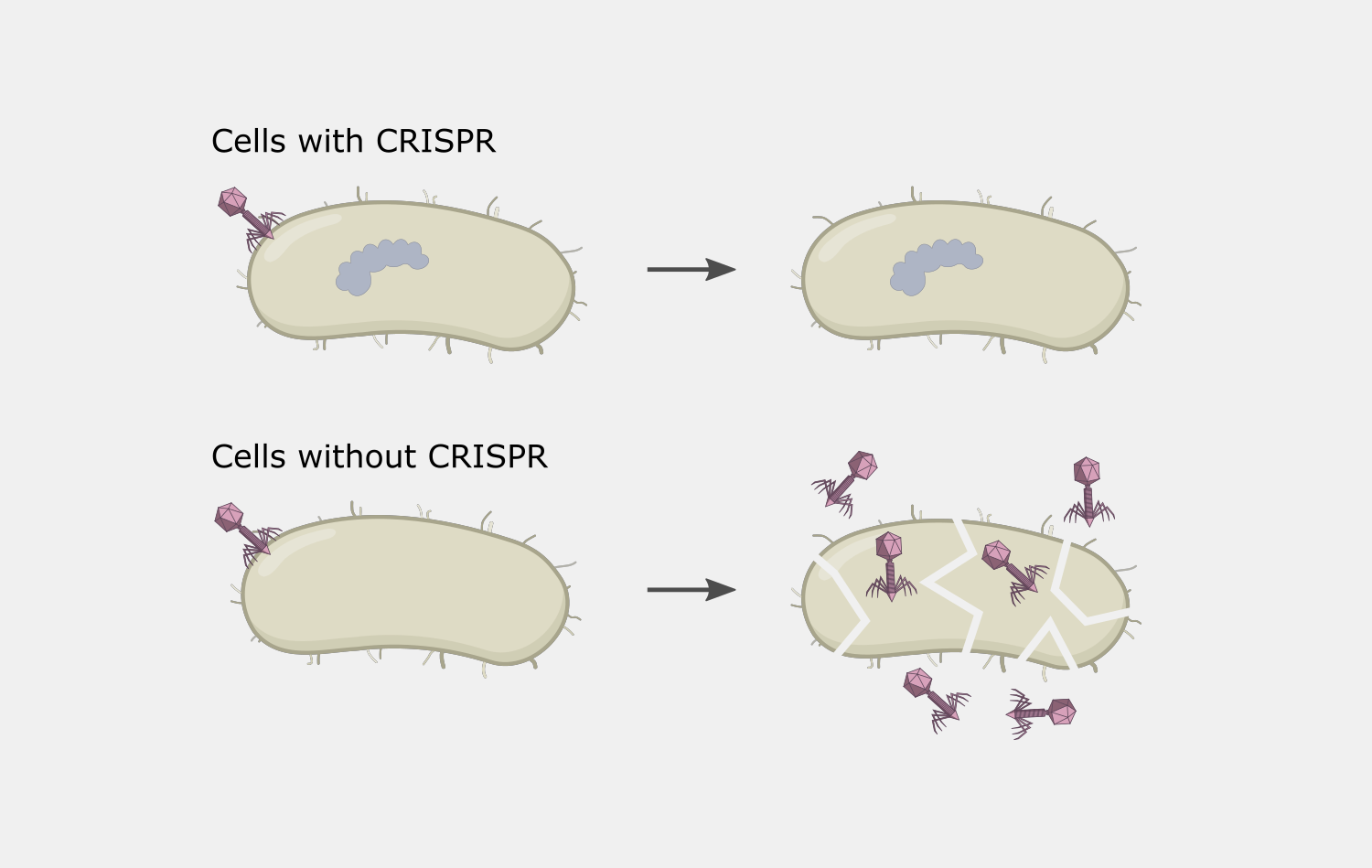
CRISPR-Cas9 gene editing tools took off rapidly because of their simplicity, flexibility and precision, characteristics that are rooted in the basic biology of the Type II CRISPR systems. Early steps in taking advantage of these characteristics to develop CRISPR-Cas9 into genome editing tools was the focus of the 2020 Chemistry Nobel Prize, awarded to Emmanuelle Charpentier and Jennifer Doudna.
Simplicity and Flexibility
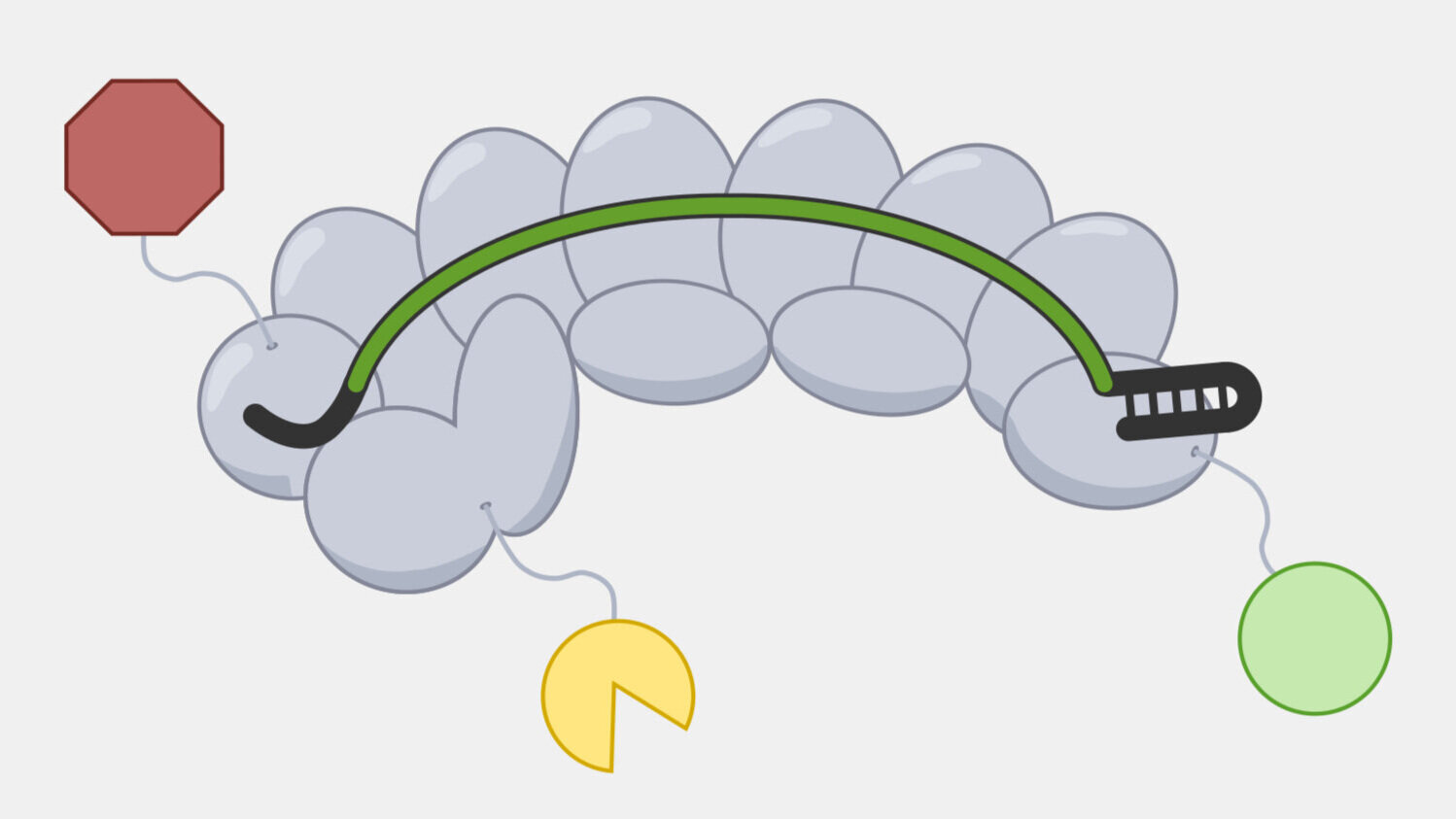
CRISPR-Cas9 system interference only requires three components: the Cas9 protein, a crRNA containing the spacer sequence for targeting, and a trans-activating crRNA (tracrRNA), making it relatively easy to deliver into many cell types, from bacteria to archaea to eukaryotes. Only he the crRNA sequence needs to be changed to redirect the complex to a new sequence, and the crRNA can be fused to tracrRNA, forming a single guide RNA (sgRNA), further simplifying the system.
Precision
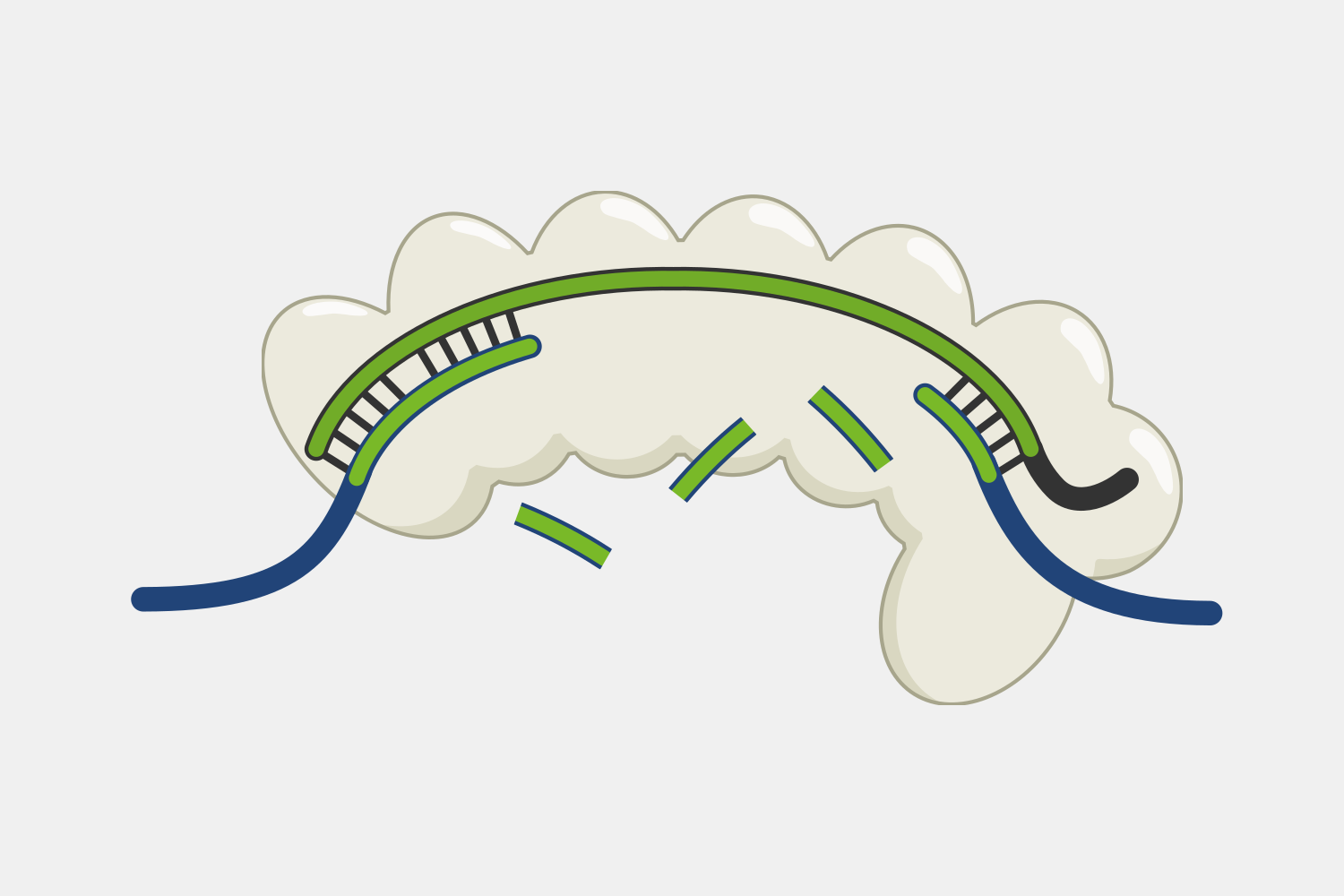
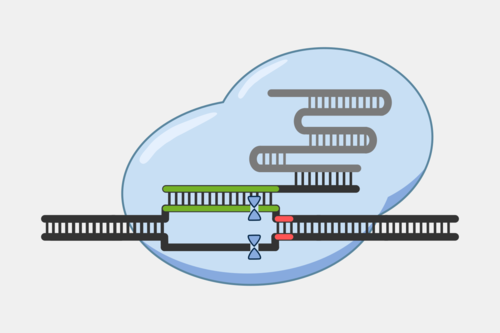
CRISPR-Cas9 systems cleave both the target and non-target DNA strand at a single location, generating a blunt cut at a specific distance from the PAM. This provides a predictable cut location, providing researchers have taken advantage of for a range of genetic engineering tools. At their most basic, Cas9 tools act as a precise pair of scissors, introducing cuts that allow for targeted deletions or insertions, often by taking advantage of DNA repair capabilities of cells, particularly non-homology end joining (NHEJ) and homology-directed repair (HDR).
Targeting
Researchers can use the programable targeting of the Cas9 complex to guide effector domains with a variety of activities beyond DNA cleavage. A Cas9 complex with the cleavage activity inactivated can still target and bind to a specific DNA sequence and will carry any fused domains along with it. For example, a Cas9 fusion to a transcriptional activator can be targeted to a region just upstream of a gene to induce transcription. Fusions with a fluorescent protein can be used to visualize the location of specific genetic sequences in a cell.
New discoveries about the basic biology of CRISPR systems continue to inform the development of CRISPR-Cas9 tools. For example, one challenge for using CRISPR-Cas9 for human gene therapy is the potential for off-target effects, when the CRISPR system alters a portion of the genome other than the target location. Further research into mechanisms of CRISPR specificity or the discovery of systems with naturally higher specificity may provide clues to reduce off-target effects in CRISPR tools.
Resources
What Is CRISPR? A brief introduction to genome editing: technology, uses, and ethics, Innovative Genomics Institute
CRISPR-Cas9 Mechanisms & Application, HHMI
The CRISPR tool kit for genome editing and beyond, Adli 2018, Nature Review
A decade of discovery: CRISPR functions and applications, Barrangou and Horvath 2017, Nature Microbiology
Breakthrough DNA Editor Born of Bacteria, Zimmer 2015, Quanta Magazine