Type III Tools
Type III systems are unique in that they cleave both RNA and DNA in a transcription-dependent manner, and some tools taking advantage of their basic biology and distribution have been created.
Programed RNA Targeting
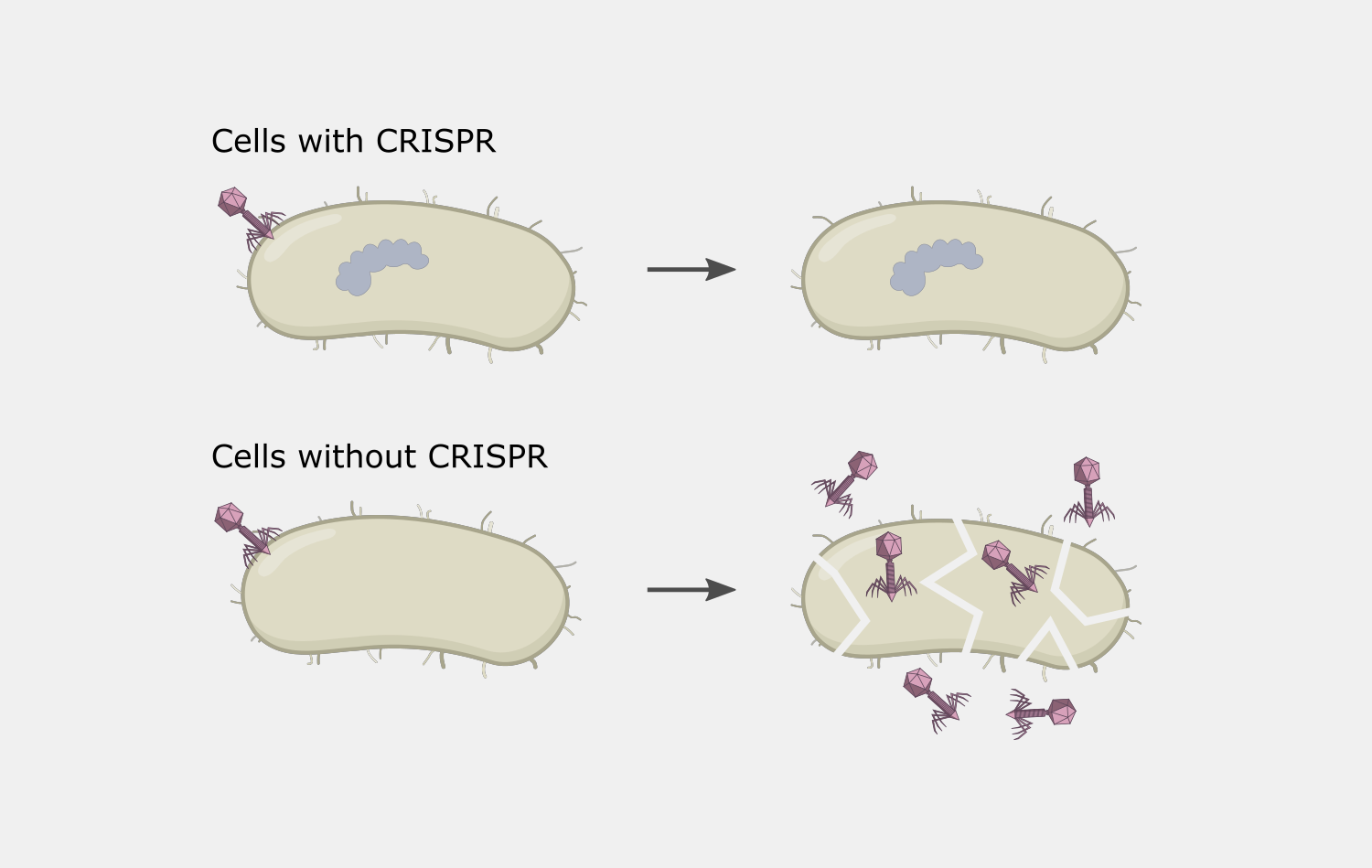
The ability of Type III systems guided by crRNA to bind and cleave RNAs provides a method to “knock-down” the levels of specific RNAs in a controlled manner when the DNA cleavage activities are disabled. Programed degradation of RNA has been demonstrated in cells with their own Type III systems by providing crRNAs. Delivery and expression of Type III system components in cells that do not have a Type III system could be used in a similar way.
Amplified RNA Cleavage
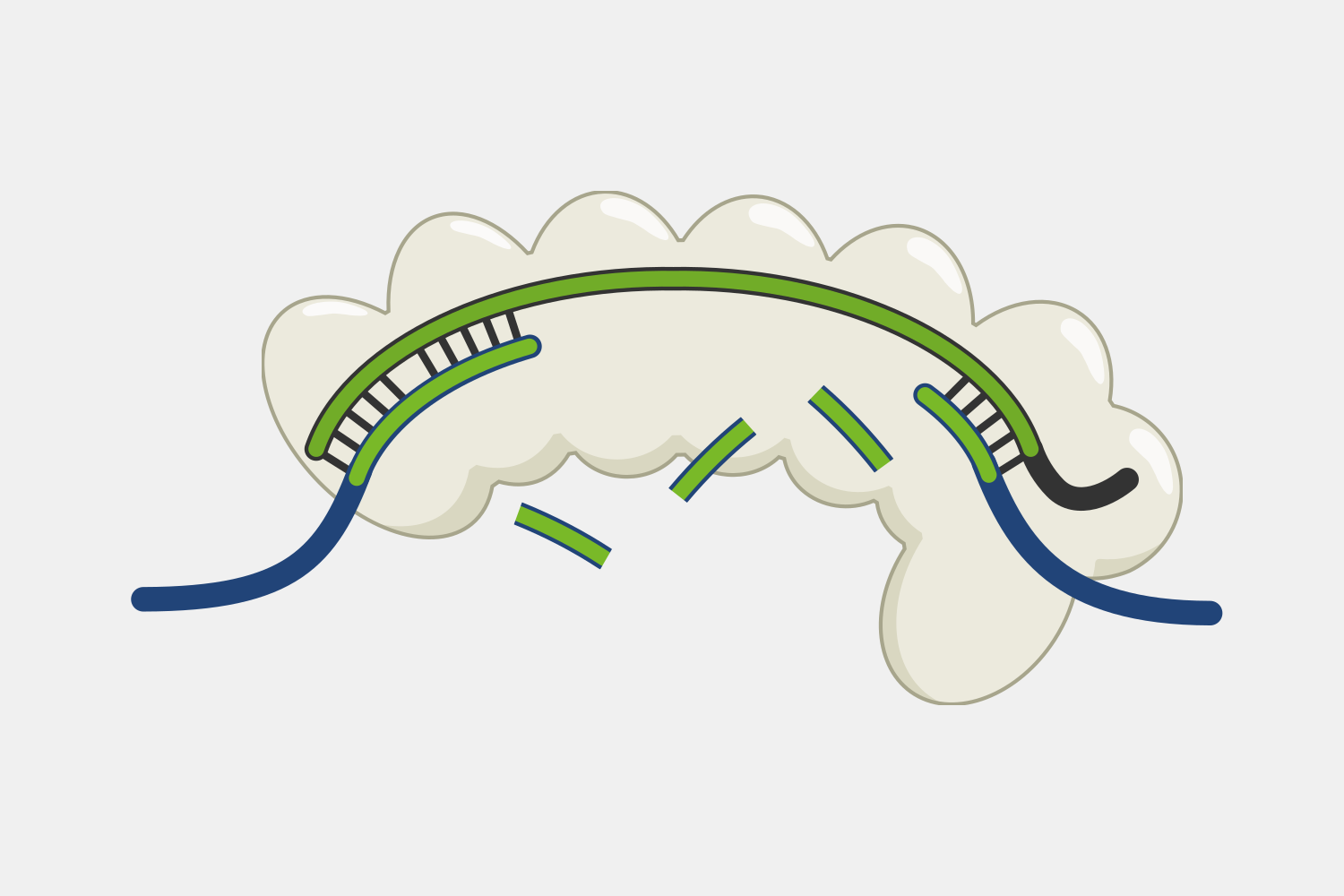
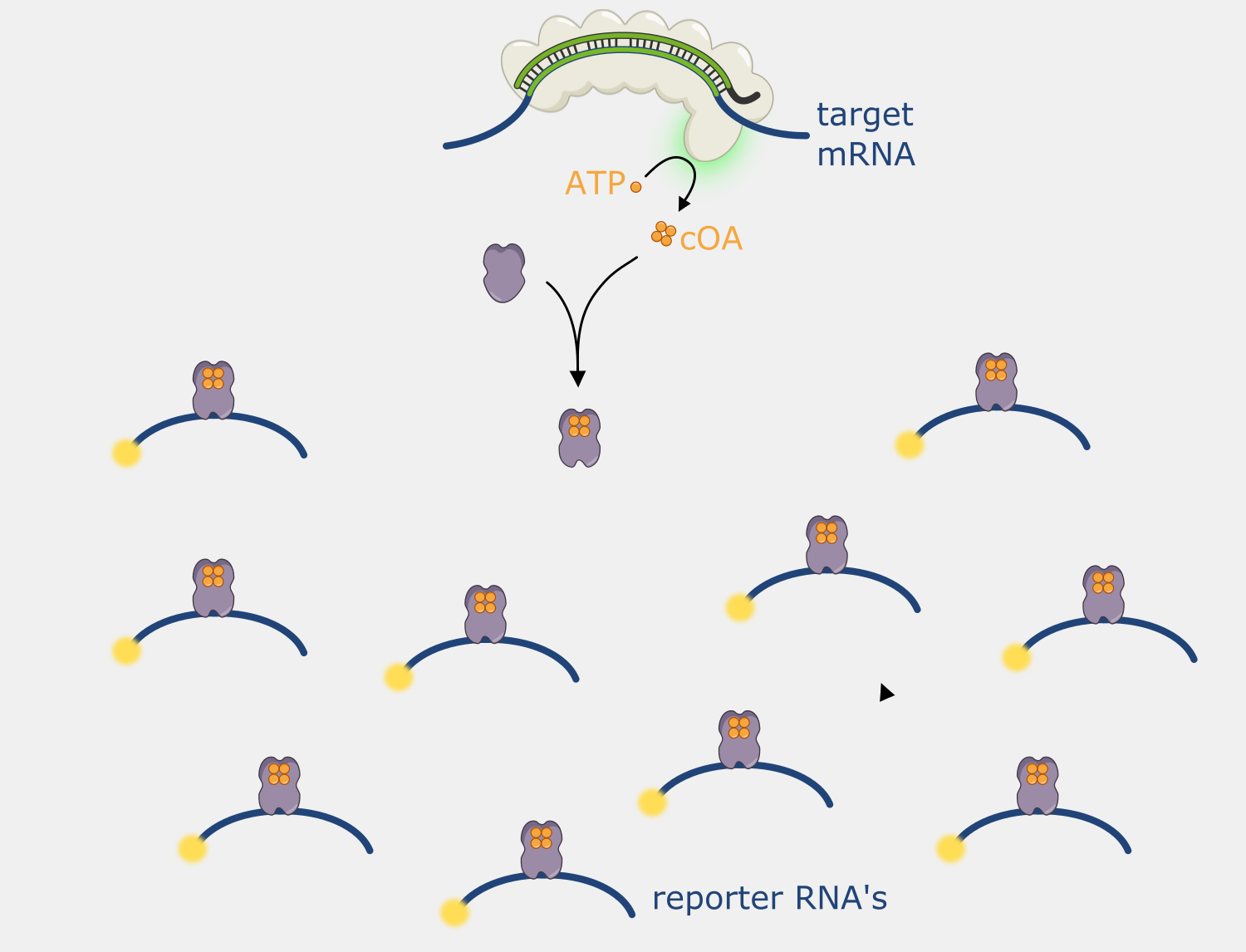
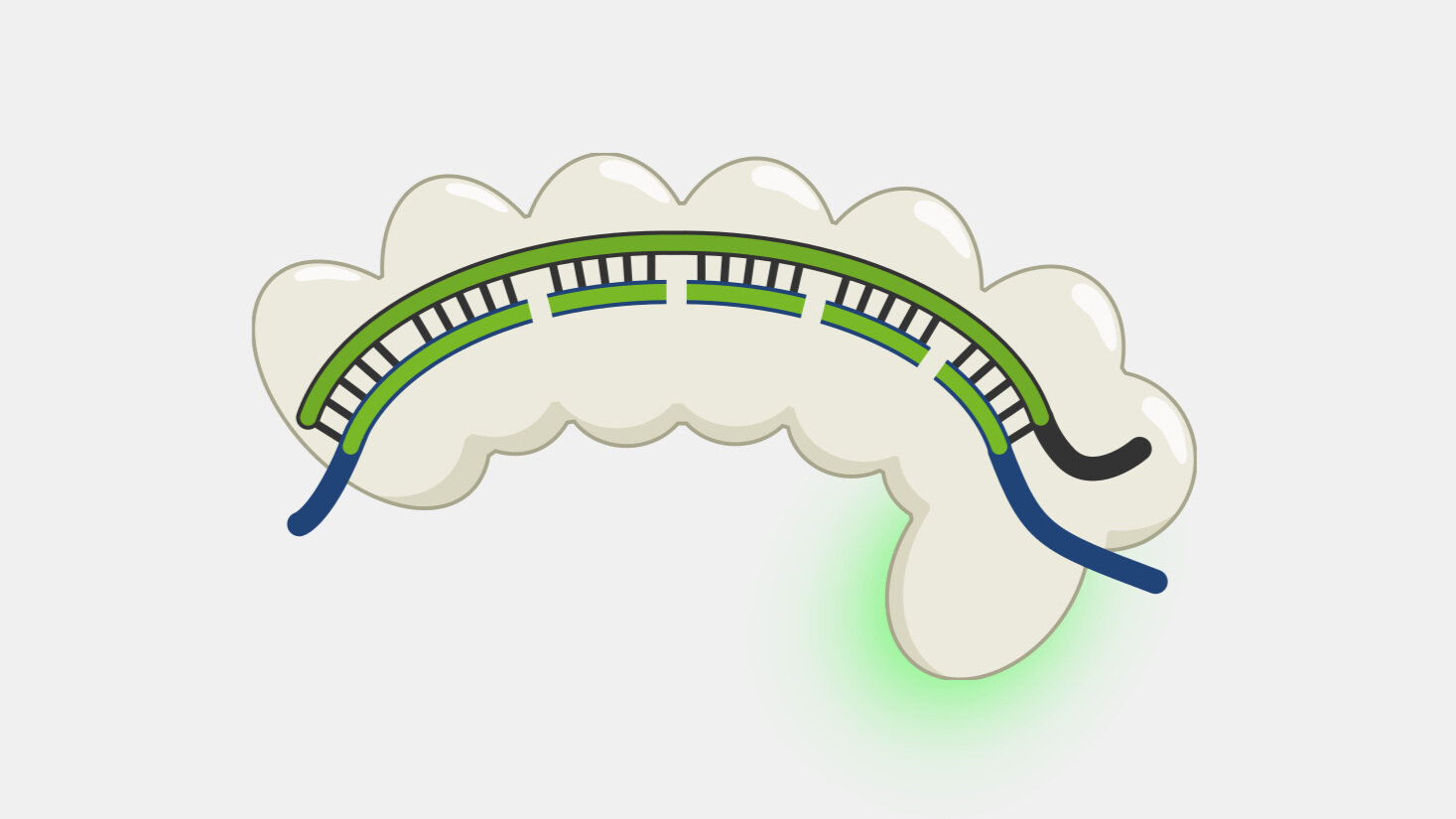
When Type III CRISPR complexes are bound to a transcript they produce cyclic oligoadenylates (cOA), which are secondary messengers that activate the RNA cleavage activity of Csm6, and each new cOA produced while the complex is bound amplifies the amount of RNA cleavage. This activity can be coupled to other systems to amplify RNA cleavage. This feature been used in a nucleic acid detection system called SHERLOCK V2, where the readout signal is based on the cleavage of RNA reporters - when the detector protein binds a target sequence, it begins cleaving the RNA reporter, triggering Csm6 to amplify the RNA cleavage signal output.
Cell Death Pathway
The potential for cyclic oligoadenylate production to trigger high enough levels of Csm6 RNA cleavage to cause extensive RNA damage leading to death is a disadvantage for some tools, but can also be used as a highly-selective antimicrobial tool. crRNAs can be created that are specific to a pathogenic species in population, or even to a particular gene, such as an antibiotic resistance gene. Type III systems only target actively transcribed genes, providing a level of control, as only cells actively expressing the genes would be affected.
The development of Type III system tools for molecular biology is currently limited, but further research into the basic biology may address some of the barriers, such as the large complexes that make delivery into cells difficult. A promising area is to develop tools that take advantage of existing Type III systems in species that have limited genetic engineering tools, but key details such as the crRNA processing and matching requirements have only been characterized for a limited number of species.
Resources
RNA-Targeting CRISPR–Cas Systems and Their Applications, Burmistrz, Krakowski and Krawczyk-Balska 2020, International Journal of Molecular Sciences
Endogenous CRISPR-Cas System-Based Genome Editing and Antimicrobials: Review and Prospects, Li and Peng 2019, Frontiers in Microbiology
CRISPR-Based Technologies: Impact of RNA-Targeting Systems, Terns 2018, Molecular Cell
Harnessing “A Billion Years of Experimentation”: The Ongoing Exploration and Exploitation of CRISPR–Cas Immune Systems, Klompe and Sternberg 2018, The CRISPR Journal