Type I Tools
Type I CRISPR systems were the first CRISPR systems to be studied, but several characteristics made them less amenable than CRISPR-Cas9 systems for certain gene editing tools. This includes their larger, multi-subunit complexes and their cleavage process, which does not result in cleanly defined blunt cut of Cas9 systems. However, some researchers are taking advantage of other aspects of Type I system basic biology to develop molecular biology tools.
Abundance
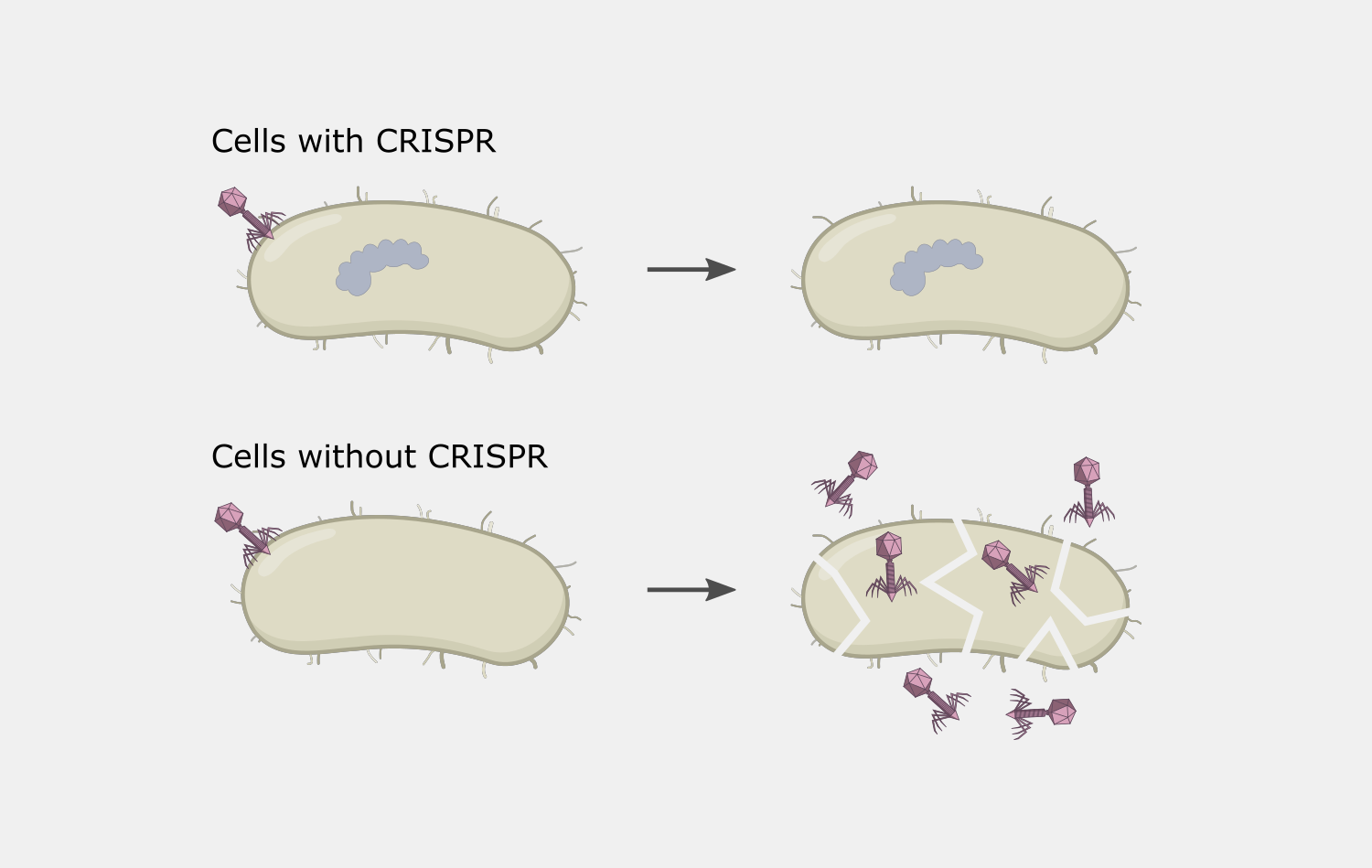
Type I CRISPR systems are the most abundant in nature, found in a large number of both bacterial and archaeal genomes, including strains of interest to humans – from pathogens to industrially useful strains involved in food and drug production. This raises the possibility of “reprogramming” cells with an endogenous Type I systems by simply providing a new crRNA. This could have several applications. A crRNA that targets the genome can result in cell death, potentially useful as antimicrobial tool. crRNAs delivered along with a repair template could be used for gene editing.
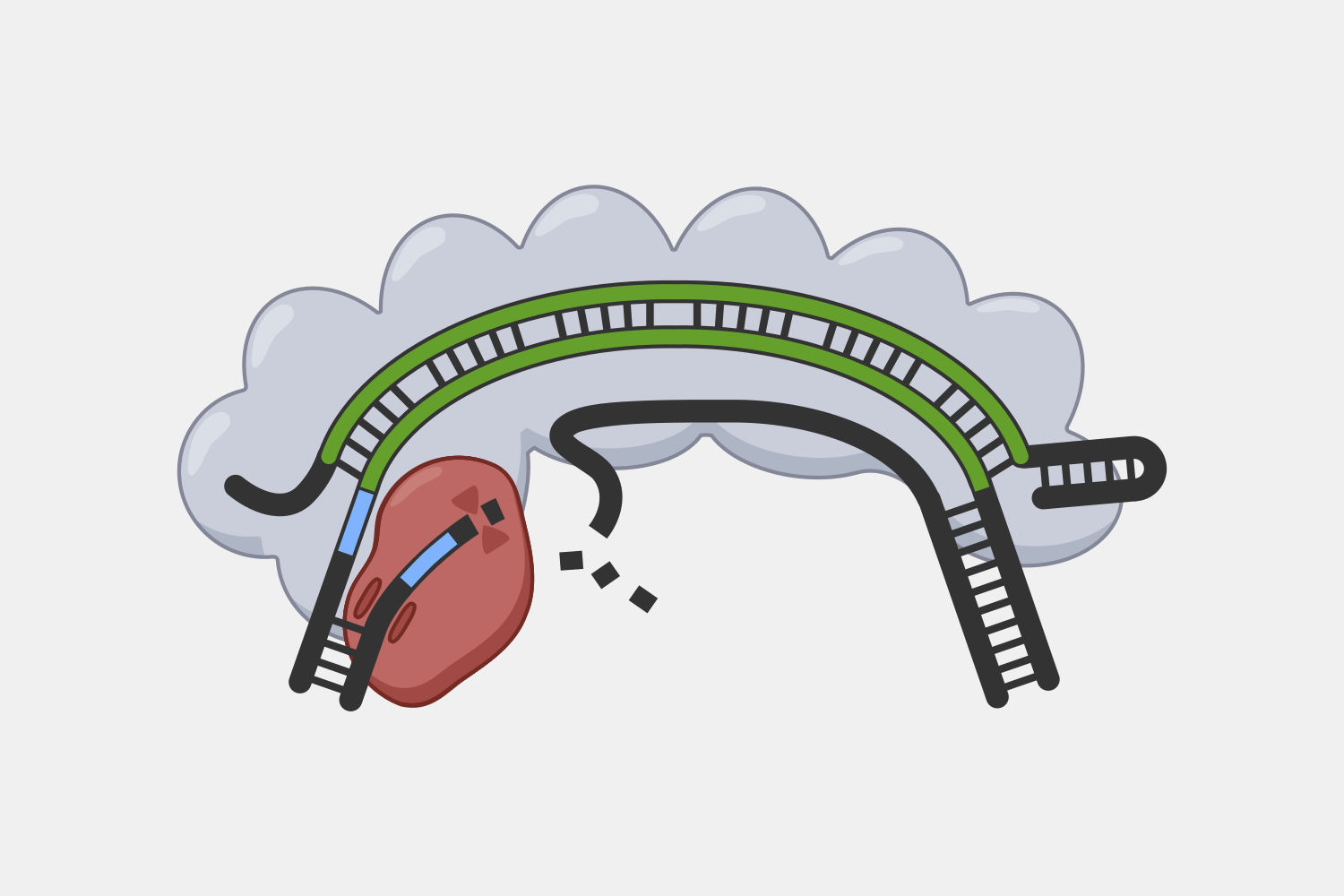
Processive Cleavage
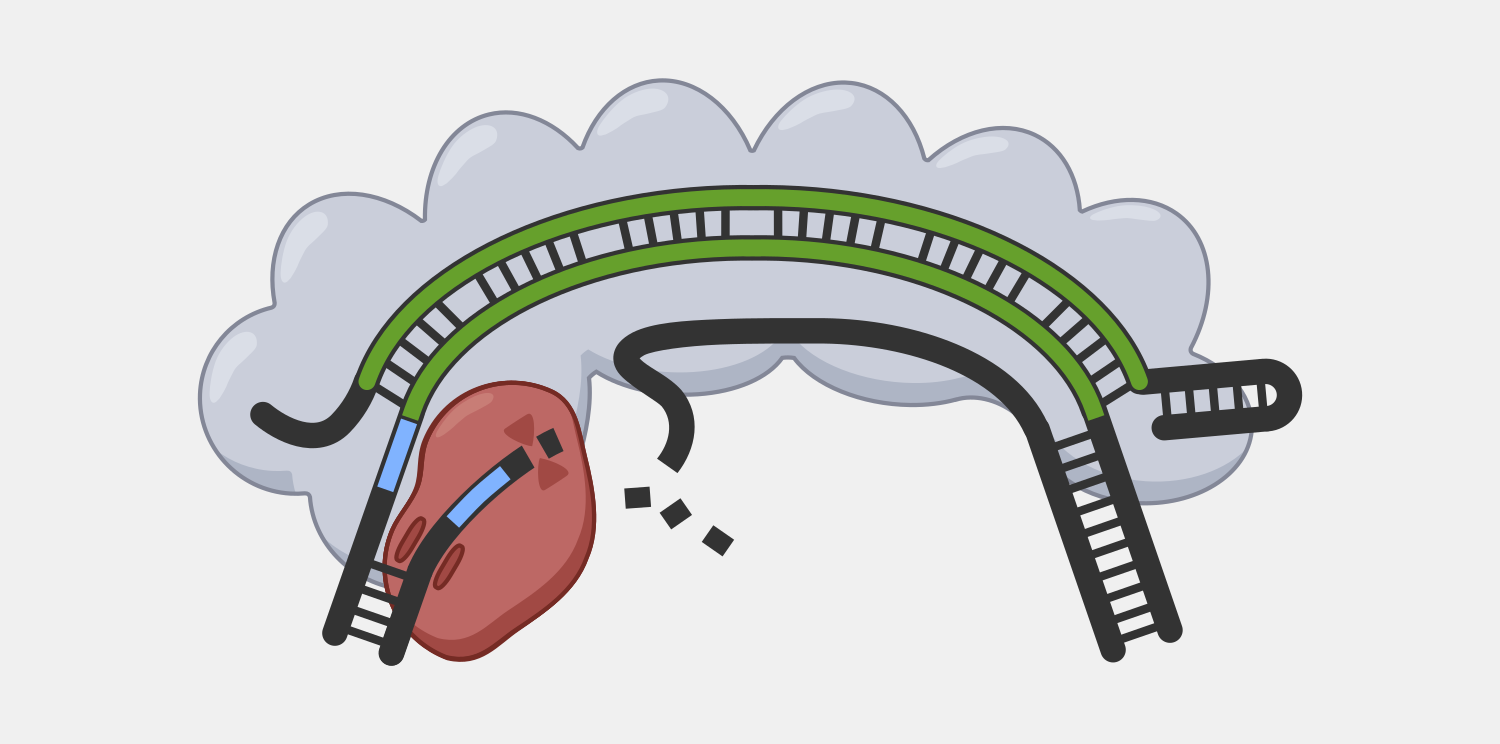
Cas3 is has both nuclease and helicase activities, which allows it to unwind double-stranded DNA and degrade the non-target strand in a 3’ to 5’ direction. While this is a problem for precise cuts, researchers have begun to take advantage of this capability to introduce large deletions in eukaryotic and prokaryotic genomes.
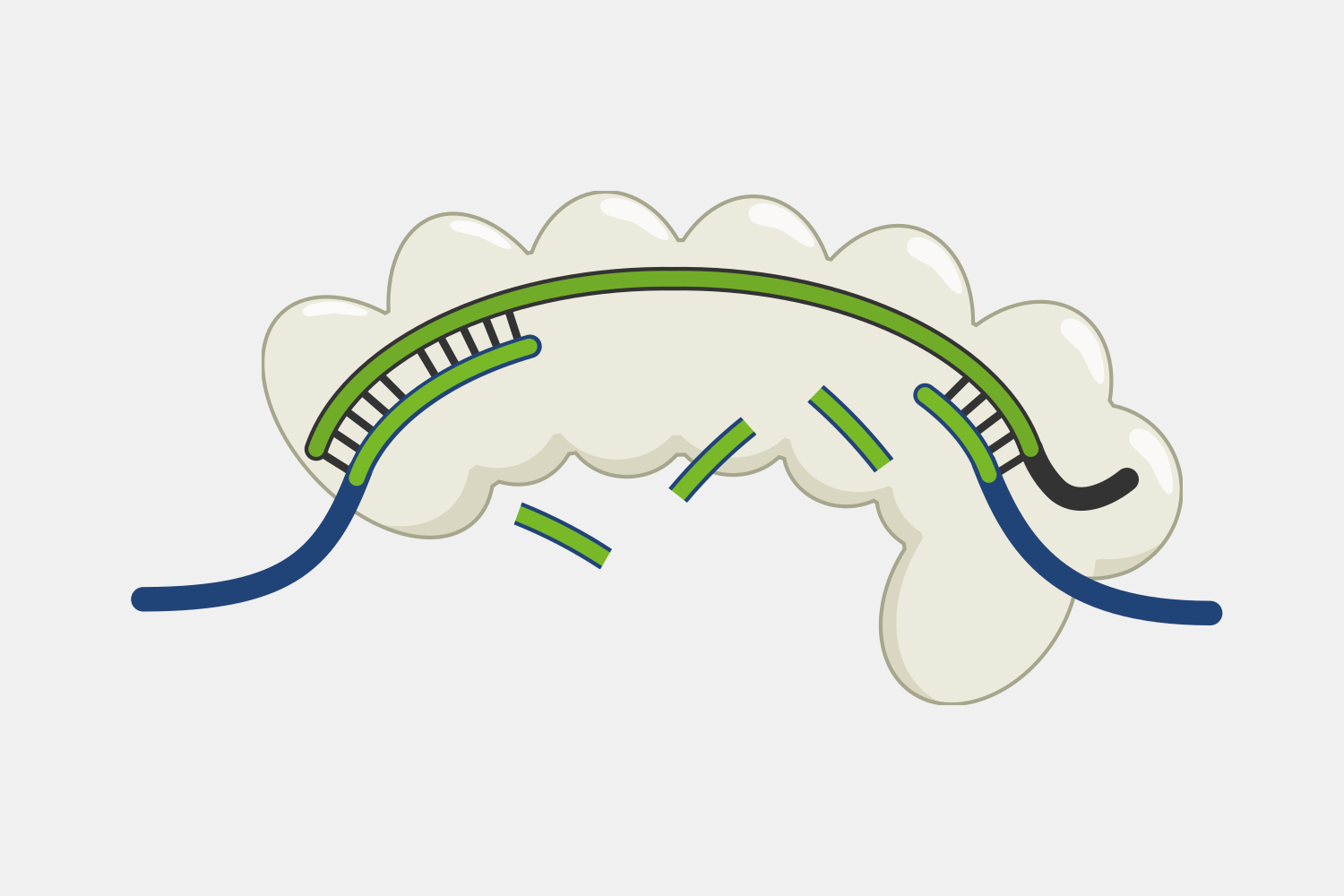
Subunit Modules
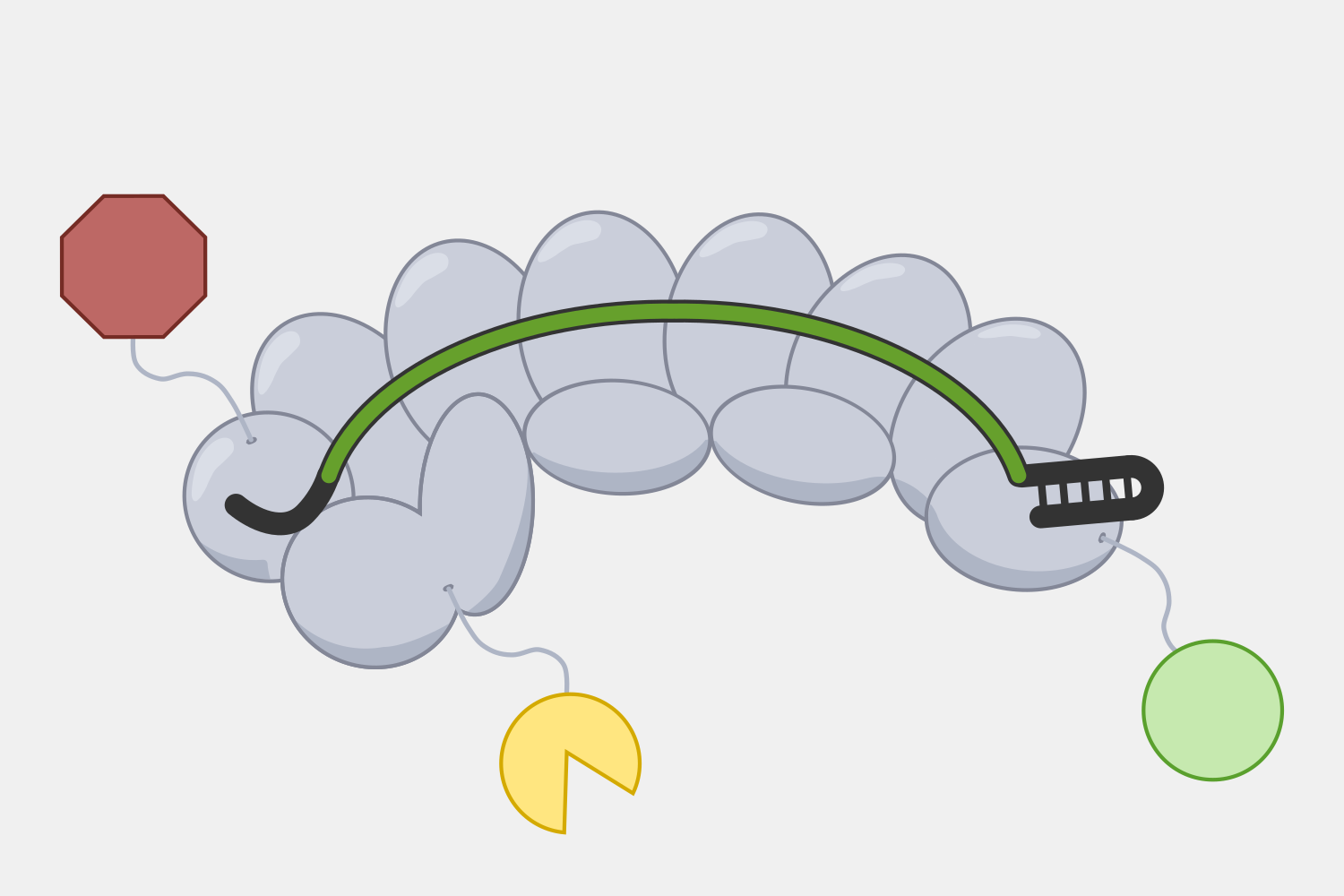
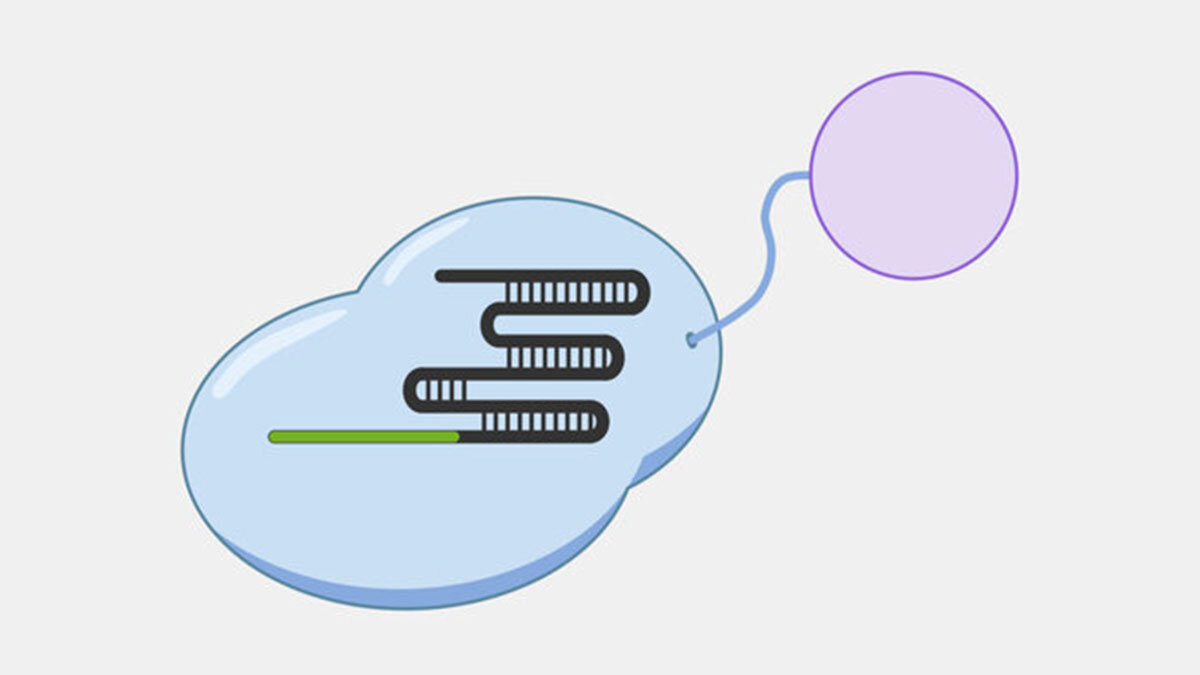
Cascade’s multiple subunits and relatively large size makes it unsuited for applications that require streamlined systems, but that modularity may offer more opportunities to engineer additional functions by modifying domains or attaching proteins with useful activities. For example, adding transcriptional activators and repressors could be used for targeted gene expression regulation, while adding proteins that make DNA alterations could be used for targeted genetic and epigenetic editing. For example, Cascade-FokI endonuclease fusions have been used to generate double stranded breaks for gene editing applications.
Taking full advantage of Type I systems for molecular biology tools will rely on further research into the basic biology of these systems. Though Type I systems are abundant in nature, only a small portion of them have been characterized. Tools that take advantage of the wide-spread endogenous systems will need to take into account the specifics of the endogenous system, such as the specific PAM sequence. Studying a broader range of these diverse systems may also uncover homologues with desirable characteristics for different applications.
Resources
Endogenous Type I CRISPR-Cas: From Foreign DNA Defense to Prokaryotic Engineering, Zheng et al, 2020, Frontiers in Bioengineering and Biotechnology
Characterization and applications of Type I CRISPR-Cas systems, Hidalgo-Cantabrana and Barrangou 2020, Biochemical Society Transactions
Characterization and Repurposing of Type I and Type II CRISPR–Cas Systems in Bacteria, Hidalgo-Cantabrana, Goh and Barrangou 2018, Journal of Molecular Biology
Snip vs Shred, Gelfand 2015, JHSPH Magazine