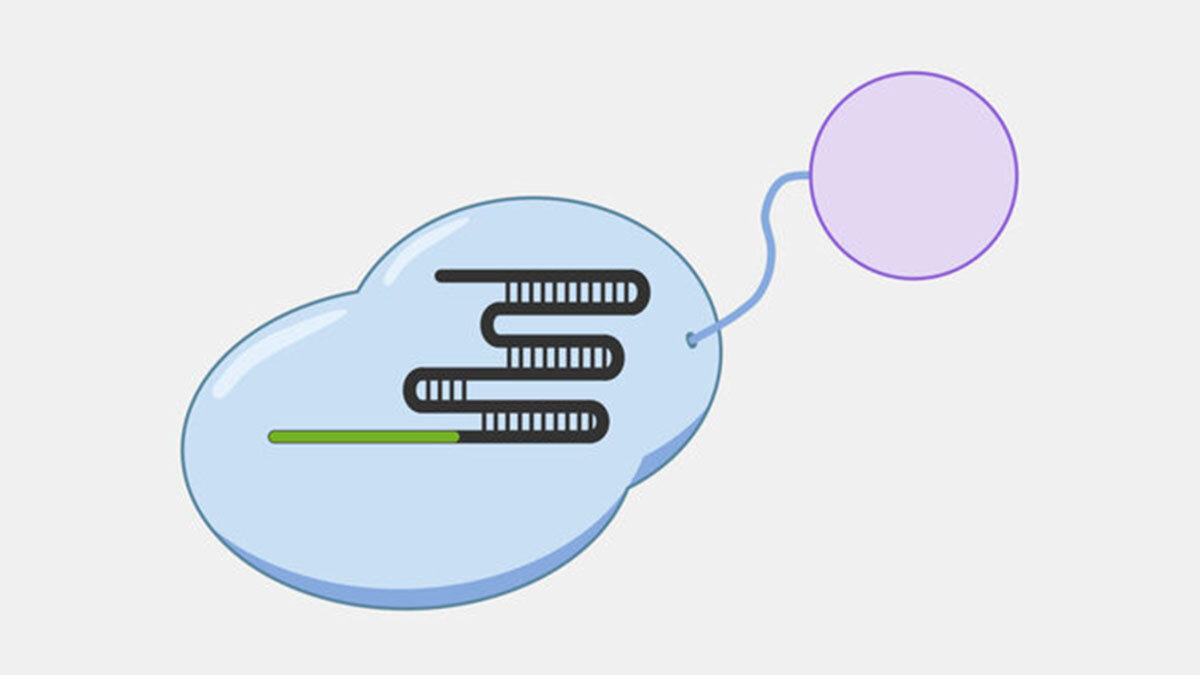
Type II Interference
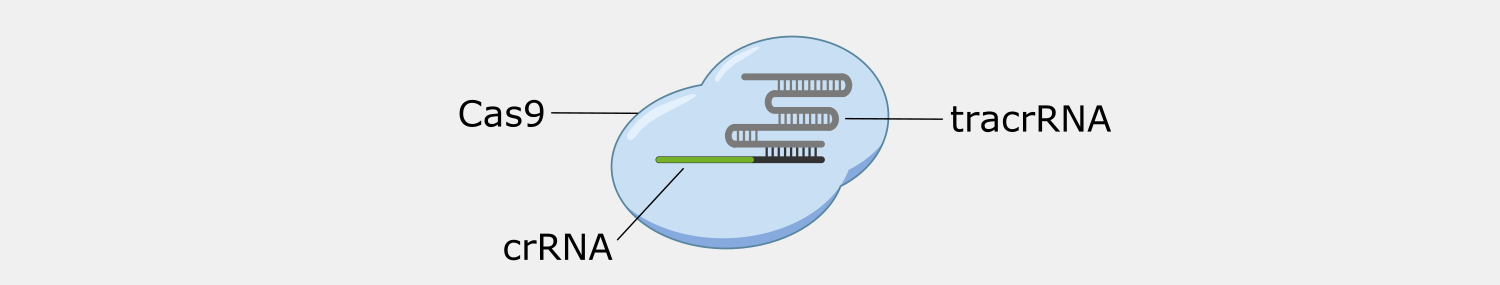
Type II systems, also known as CRISPR-Cas9 systems, are widely-known because of their use as genetic engineering tools, which was the subject of the recent Nobel Prize in Chemistry, awarded to awarded to Emmanuelle Charpentier and Jennifer Doudna. Type II systems identify and cleave double-stranded DNA from phages, and use a CRISPR complex containing a single protein, Cas9, and two RNAs. In addition to the crRNA found in all CRISPR complexes, Type II complexes have a tracrRNA (trans-activating crRNA), a small RNA that plays a role in the assembly and activation of the Cas9 complex and includes a region that complements a portion of the crRNA. The Cas9 complex both recognizes and cleaves the double-stranded DNA targets.
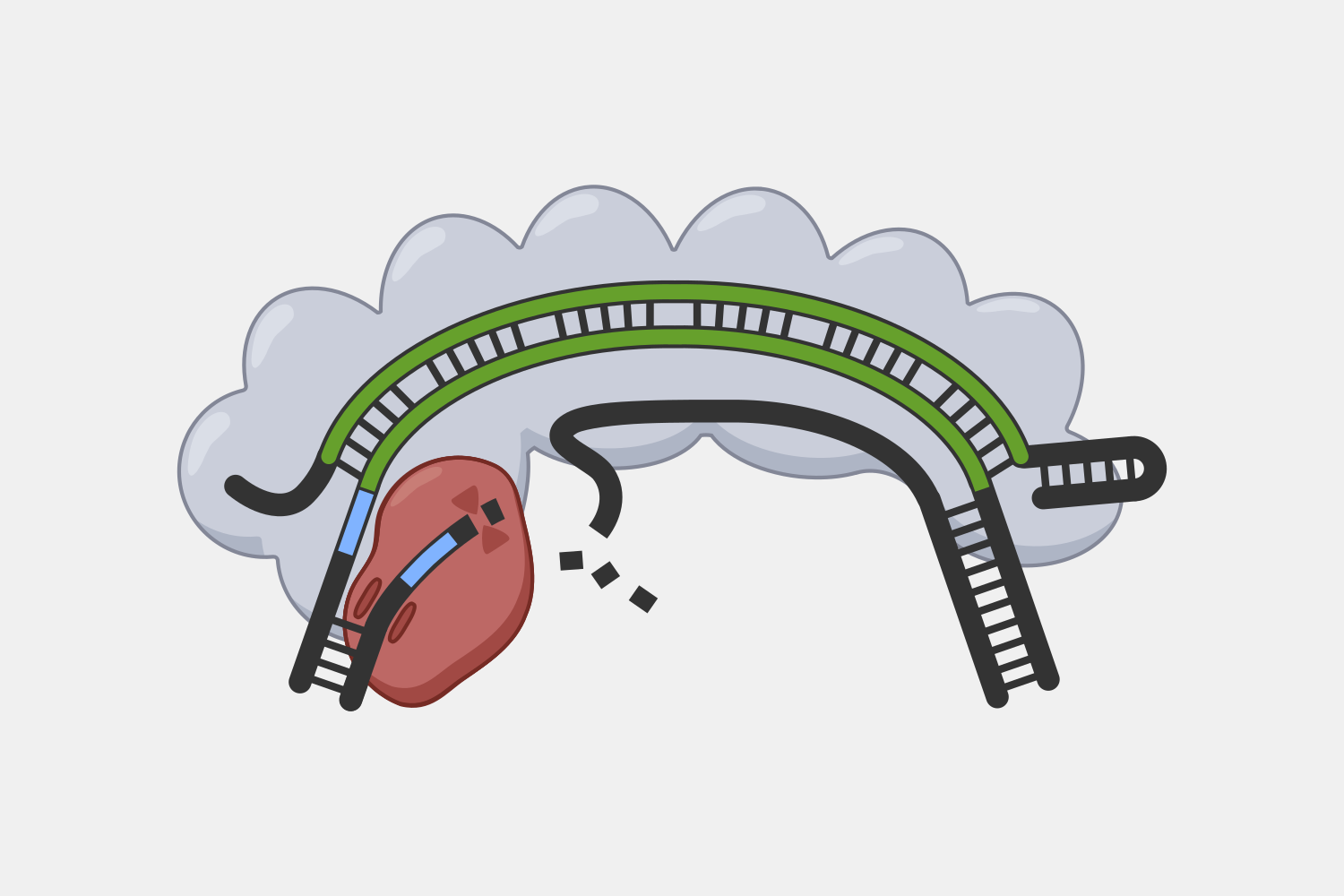
Cas9 complexes target DNA sequences that match their crRNAs, but cells are filled with double-stranded DNA, and sequences that match the crRNA are rare – if they are even present. Unwinding all that DNA to check for regions complementary to the crRNA spacer would not be efficient. Instead, the Cas9 complex searches for protospacer adjacent motif (PAM) sequences in double-stranded DNA.
In the original foreign DNA fragment used to generate the CRISPR array, and the PAM is found next to the protospacer, the sequence that is added as a CRISPR array spacer. The Cas9 complex identities the PAM without needing to first unwind the DNA. Though the PAM sequence is not unique to the phage DNA target, only searching near a PAM significantly reduces the number of locations to unwind.
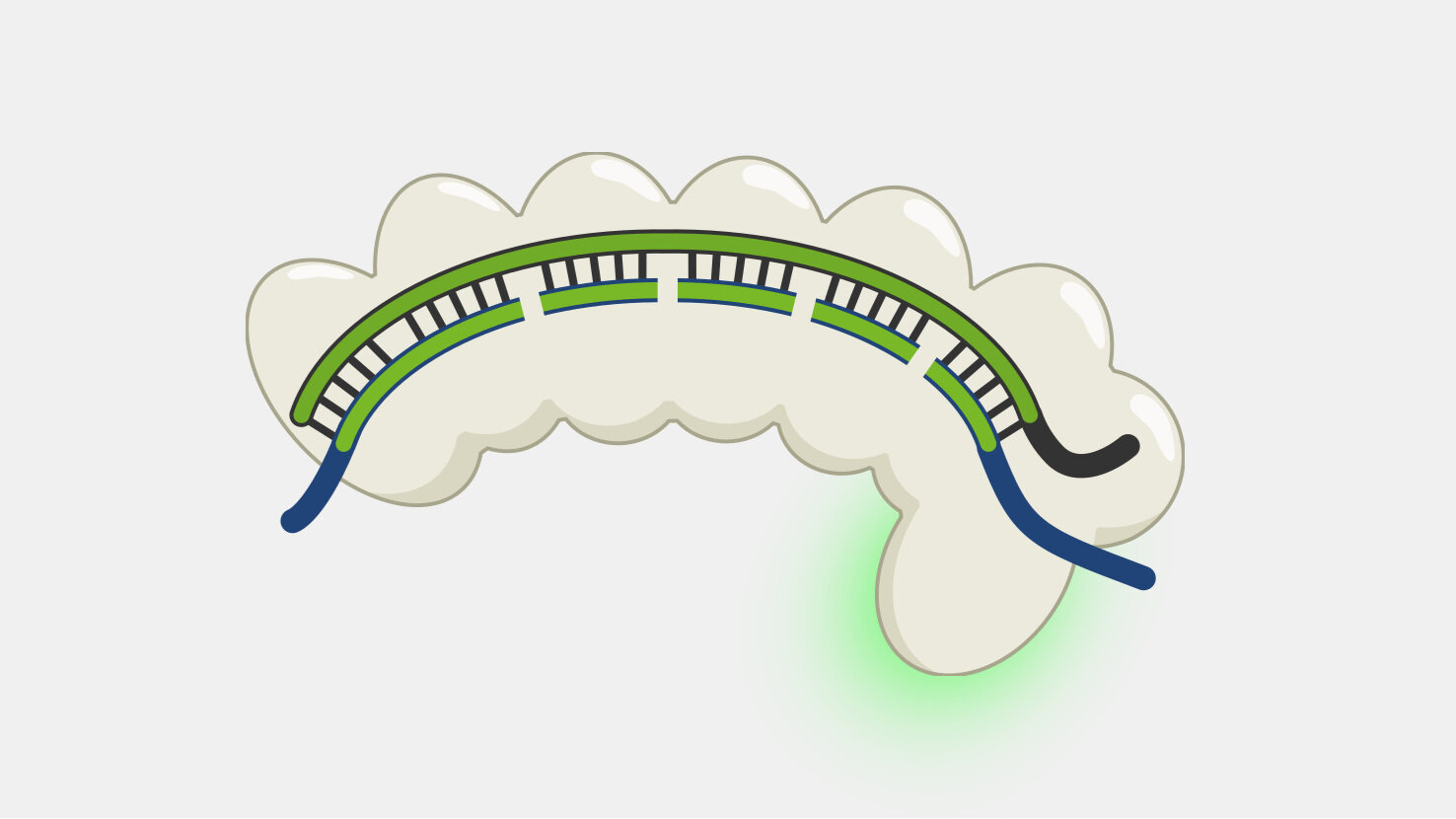
When the Cas9 complex recognizes a PAM sequence it binds and begins to unwind the double-stranded DNA, exposing the target strand and allowing the crRNA bases to bind complementary sequences in the target strand.
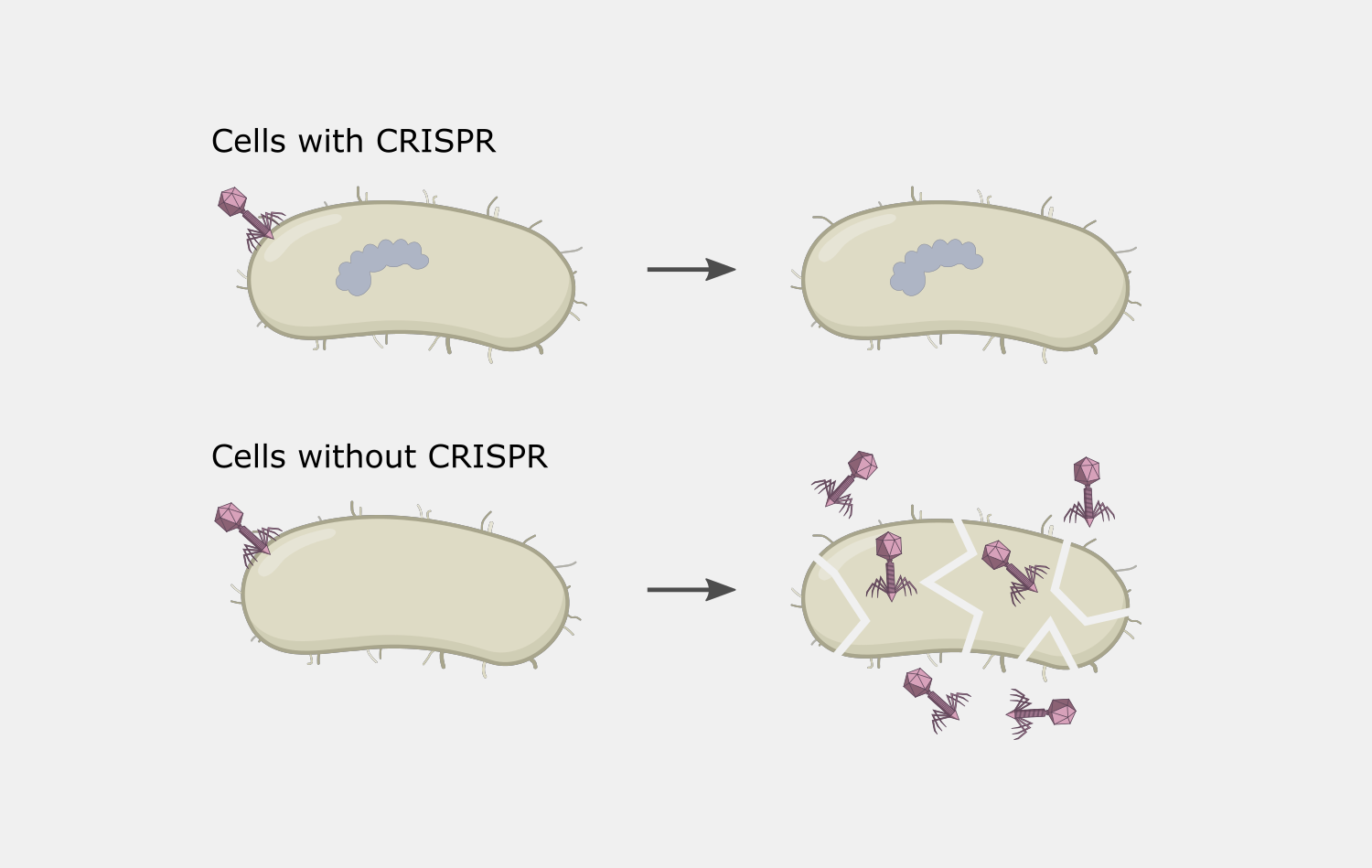
In Type II systems, the pairing of the target strand to the crRNA activates two nucleases domains, which each cleave one strand of the DNA in the same location, resulting in a blunt cut. This activity results in destruction of the DNA target, blocking the phage infection and providing immunity.
CRISPR interference could cause autoimmunity if the complex was to target and cleave the spacer sequence in the host’s CRISPR array. In Type II systems, the PAM binding requirement reduces the chances of this, because the PAM is not part of the spacer sequence found in the CRISPR array, so the Cas9 complex will not bind to the array.
Resources
Related Bailey Lab Research
Haobo Wang, John Mallon and Scott Bailey collaborated on research lead by Taekjip Ha’s lab that analyzed the DNA unwinding activity of Cas9 and four engineered versions on DNA targets with and without mismatches, using different variations of guide RNA (a single RNA that combines the crRNA and tracrRNA). The results provided insights into the unwinding process and how different aspects are affected by mismatches. Single molecule analysis of effects of non-canonical guide RNAs and specificity-enhancing mutations on Cas9-induced DNA unwinding. Okafor IC, Singh D, Wang Y, Jung M, Wang H, Mallon J, Bailey S, Lee JK, Ha T. (2019) Nucleic Acids Res. 47(22):11880-11888.
John Mallon and Scott Bailey collaborated on research lead by Taekjip Ha’s lab using single-molecule imaging to examine how engineered versions of Cas9s interact with DNA targets containing sequence mismatches, focusing on the surveillance and unwinding steps. The team found that mismatches nearest the PAM impaired stable binding the most, while mismatches in the region furthest from the PAM impaired DNA unwinding. They also showed that cleavage followed the unwinding of the DNA, and not at the same time as, or because of, the nuclease activity. Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis. Singh D, Wang Y, Mallon J, Yang O, Fei J, Poddar A, Ceylan D, Bailey S, Ha T (2018) Nat. Struct. Mol. Biol. 25: 347-354.
Hongfan Chen, Jihoon Choi and Scott Bailey characterized the Streptococcus thermophilus LMG18311 Cas9 system’s PAM and DNA cleavage requirements. Using in vitro experiments, they also showed that the two Cas9 nuclease domains use have different cleavage site selection methods: the HNH domain cuts the target strand at a fixed position within to the region complementary to the crRNA, while the RuvC-like domain cuts the non-target strand using a ruler mechanism, based on a distance from the PAM sequence. Independent Cut Site Selection by the Two Nuclease Domains of the Cas9 RNA-guided Endoribonuclease. Chen H, Choi J, Bailey S (2014) J. Biol. Chem. 289: 13284-13294. PDF
Reviews
Harnessing “A Billion Years of Experimentation”: The Ongoing Exploration and Exploitation of CRISPR–Cas Immune Systems, Klompe and Sterberg, 2018. The CRISPR Journal
The Biology of CRISPR-Cas: Backward and Forward, Hille et al, 2018. Cell